PCA (HD)
Uses an orthogonal transformation to convert a set of observations of possibly correlated variables into a set of values of uncorrelated variables (principal components).

Information at a Glance
|
Parameter |
Description |
|---|---|
| Category | Model |
| Data source type | HD |
| Send output to other operators | Yes |
| Data processing tool | In-memory: MapReduce or Spark, depending on input configuration |
Algorithm
PCA (Principal Component Analysis) is an orthogonal linear transformation that transforms data into a new coordinate system such that the greatest variance by any projection of the data comes to lie on the first coordinate (called the first principal component), the second greatest variance on the second coordinate, the third on the third coordinate, continuing until the number of coordinates has been reached or a preset maximum principal component threshold has been reached.
The Alpine PCA operator implements an eigenvalue decomposition of a data covariance matrix Σ (or, correlation matrix R).
- Each principal component is a linear combination of the original variables.
- The coefficients (loadings) are the eigenvectors (v1, v2,...vp) of covariance matrix Σ (or, correlation matrix R) with unit length.
- The eigenvalues (λ1, λ2,...λp) denote the contribution of the principal component associated with it.
- The principal components are sorted by descending order according to their variance contribution.
- The user can choose the number of principal components according to the accumulation contribution (∑ij=1λj/∑pK=1λK).
More details are available in Principal Component Analysis, (1986), Joliffe, I.T.
Additional references:
- Jerome Friedman, Trevor Hastie, Robert Tibshirani (2008), The Elements of Statistical Learning Data Mining, Inference and Prediction Chapter 3: "Linear Methods for Regression"
- Joliffe, I.T. (1986), Principal Component Analysis, New York, Springer
- Wu, W., Massart, D.L., and de Jong, S. (1997), "The Kernel PCA Algorithms for Wide Data. Part I: Theory and Algorithms" Chemometrics and Intelligent Laboratory Systems, 36, 165-172.
Input
A data set from the preceding operator.
Configuration
| Parameter | Description |
|---|---|
| Notes | Notes or helpful information about this operator's parameter settings. When you enter content in the Notes field, a yellow asterisk appears on the operator. |
| Columns | Click Columns to open the dialog for selecting the available variable columns to transform by the PCA algorithm, in order to create a reduced set of variables. |
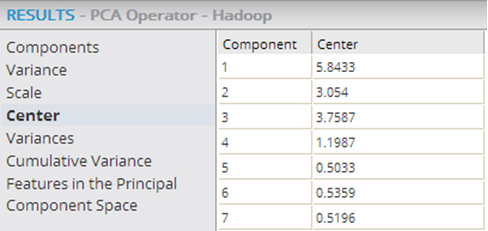
| Center | If
Yes (the default), the mean value of each variable column is set to 0 before the PCA matrix transformation algorithm is run. In combination with
Scale, the following applies.
Usually, the PCA algorithm has meaning only if the data is centered first, so the default is set to Yes. When Use Spark is set to Yes, then this parameter is grayed out. |
| Scale | When the
Scale option is selected, each variable's data values are divided by the Standard Deviation so that all of the columns have the same data spread (that is, to be on an equivalent scale as each other).
See Center for more information about its effect on the algorithm. Default value: Yes. Note that variables with zero Standard Deviation should not be input into PCA. When Use Spark is set to Yes, then this parameter is grayed out. |
| In Memory Threshold | Determines whether to compute the PCA by a Hadoop MapReduce job or in memory SVD (single machine instead of distributed mode).
If the number of rows in the training data set is fewer than the threshold value, the PCA algorithm is run in memory SVD. Otherwise, it is computed by a MapReduce job. When the input parameter Use Spark is set to Yes, then the job is run in Spark regardless of the value of this parameter, which is grayed out. Note: The MapReduce option is currently deprecated.
|
| Maximum Number of Components to output. | Determines the upper-limit number of principal components to calculate, starting with the top, high-ranking variance components. This value must be equal to or typically less than the number of columns in the training data set, to help with dimension reduction.
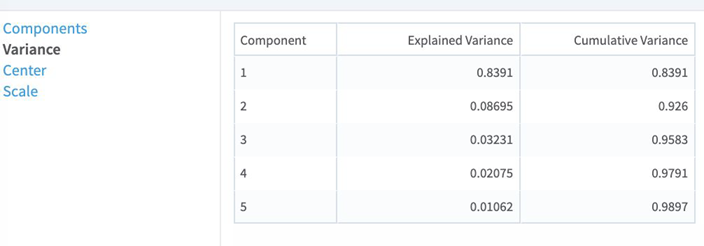
The choice of this value depends on the desired cumulative variance to cover. To check whether this number would return enough components, inspect the output tab Variance (when Use Spark is set to Yes) or the output tab Cumulative Variance (when Use Spark is set to No). |
| Additional Runs for Distributed Mode | Specifies the number of required extra passes of the algorithm to implement when computing in-memory SVD. Typically, a single pass result is sufficient when the number or rows are less than the In Memory Threshold value.
Default value: 0 (no additional runs). When Use Spark is set to Yes, then this parameter is grayed out. |
| Max JVM Heap Size (MB) (-1=Automatic) | A Java Virtual Machine data storage setting for Hadoop. (Spark option only.)
When Use Spark is set to Yes, then this parameter is grayed out. |
| Use Spark | If Yes (the default), uses Spark to optimize calculation time. |
| Advanced Spark Settings Automatic Optimization |
|
Output
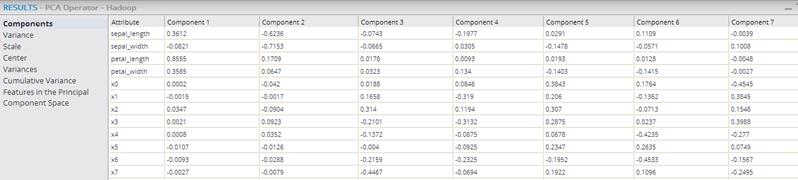
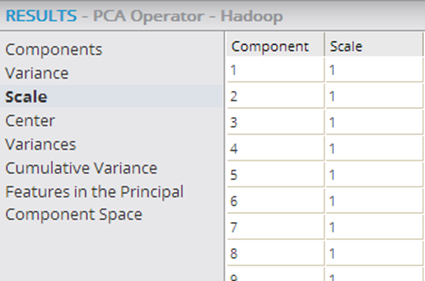
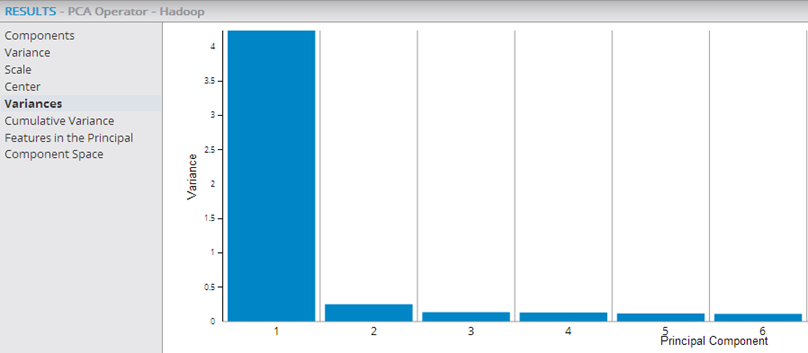
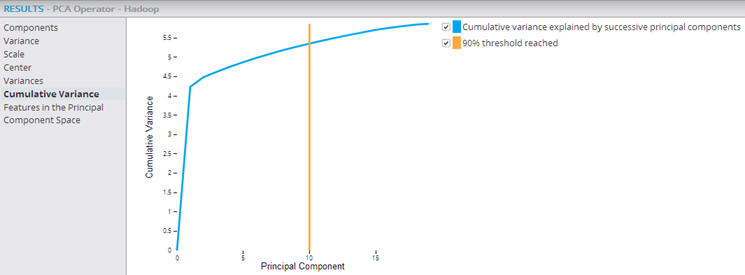
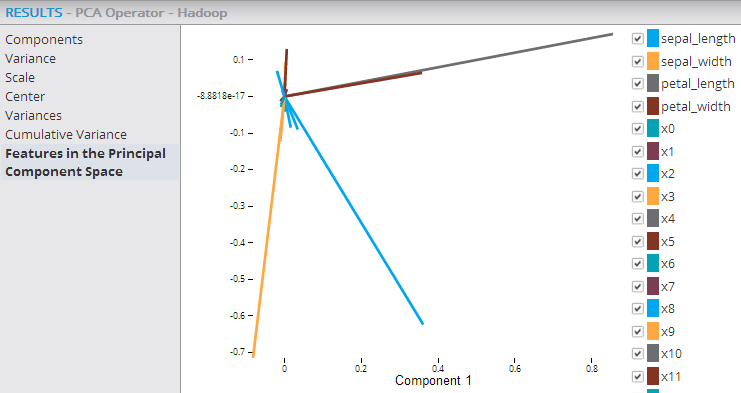
The following example shows the PCA and Predictor operators within a Hadoop workflow, with their output being passed into the Correlation operator, which confirms that the added transformed variables are uncorrelated.
See the Predictor (HD) operator for more details.
Example