SOM Clustering
This operator implements the self-organizing maps algorithm that generates the clusters spatially aligned according to their similarity.
Information at a Glance
|
Parameter |
Description |
|---|---|
| Category | Model |
| Data source type | TIBCO® Data Virtualization |
| Send output to other operators | Yes |
| Data processing tool | TIBCO® DV, Apache Spark 3.2 or later |
Algorithm
The self-organizing map clustering algorithm is a simple neural network that produces clusters spatially aligned on a grid, where clusters close to each other are more similar to each other. The columns specified are used to train the self-organizing maps clustering algorithm.
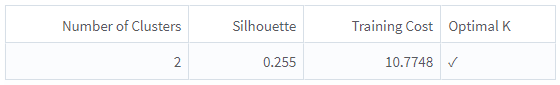
This operator uses the Silhouette value to determine the optimal number of clusters. The silhouette metric is a measure of how similar observation is to its assigned cluster compared to other clusters. Generally, the higher silhouette values are preferred.
Input
An input is a single tabular data set.
Configuration
The following table provides the configuration details for the SOM Clustering operator.
| Parameter | Description |
|---|---|
| Notes | Notes or helpful information about this operator's parameter settings. When you enter content in the Notes field, a yellow asterisk appears on the operator. |
| Use all available columns as Predictors | When set to Yes, the operator uses all the available columns as predictors and ignores the Continuous Predictors and Categorical Predictors parameters. When set to No, the user must select at least one of the Continuous or Categorical Predictors. |
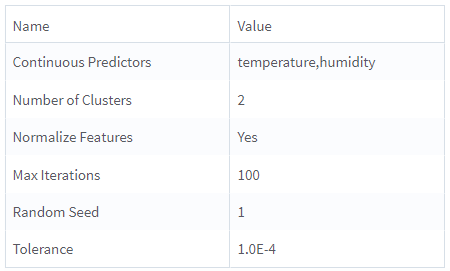
| Continuous Predictors | Specify the numerical data columns for training the self-organizing maps model. It must be numerical column. Click Select Columns to select the required columns. |
| Number of Clusters | Specify the grid dimensions, which control the number of clusters to create during self-organizing maps. The following methods are available:
Default: 2 |
| Normalize Features | Specify whether to normalize the numerical features (Z-Transformation). Default: Yes |
| Max Iterations | Specify the maximum number of iterations for one run of the self-organizing map clustering algorithm (one iteration per sample). Default: 100 |
| Tolerance | The smaller the value, the stricter the determination of when the analysis has converged. A smaller number results in more iterations of the algorithm but are capped by the iteration limit. Default: 1.0E-4 |
| Random Seed | Specify the seed used for the pseudo-random row extraction. Default: 1 |
Output
- Parameter Summary Info: Displays information about the input parameters and their current settings.
- Training Summary: A text field that displays the training summary.
Silhouette: A measure to compare the similarity of observation to its assigned cluster compared to other clusters.
Training Cost: The sum of specified distances to the nearest centroid for all points in the training data set.
-
PRED_SOM: Specifies the cluster to that an observation belongs.
-
DIST_SOM: The distance between the cluster centroid and the observation.
Example
The following example illustrates the SOM Clustering operator.
- Multiple columns namely outlook, temperature, wind, humidity, and play.
- Multiple rows (14 rows).
-
Use all available columns as Predictors: No
-
Continuous Predictors: temperature, humidity
-
Number of Clusters: 2
-
Normalize Features: Yes
-
Max Iterations: 100
-
Tolerance: 1.0E-4
-
Random Seed: 1