Naive Bayes Use Case
Naive Bayes modeling is useful for a variety of different applications in the biological and medical fields, in online document and spam classification areas, for supply chain stock management, and for financial predictions. Here is a specific use case example from the biological field.
- Datasets
-
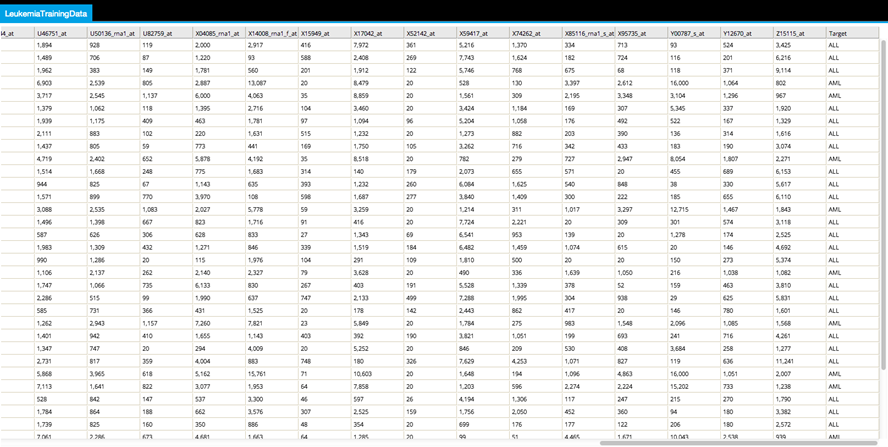
The source dataset for this use case is LeukemiaTrainingData.csv (9KB). Training biological leukemia gene dataset used in this use case was obtained from: http://research.cs.queensu.ca/home/xiao/dm.html.
This example dataset was created from biological study measurements - the data was cleaned, normalized and filtered down to the top 31 genes associated with leukemia. Then the data was transposed so that each column represents an Attribute (Gene name) and each row represents a biological sample record. The Class column was added to indicate whether the type of leukemia related to the sample was either "ALL" or "AML".
The goal of applying the Naive Bayes model on the dataset is to find those genes, or attributes, that have high discriminating ability to distinguish between different types of leukemia. Therefore, in this case, the Dependent Value represents the Type of Leukemia, which is a categorical variable with possible values of either "ALL" or "AML".
The first rows of this dataset are shown below:
Workflow
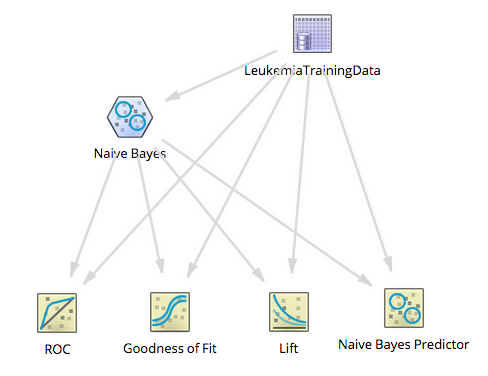
For this use case, a Naive Bayes model can be built from the training Naive Bayes Leukemia dataset by adding the Naive Bayes and Naive Bayes Prediction Operators in a new flow:
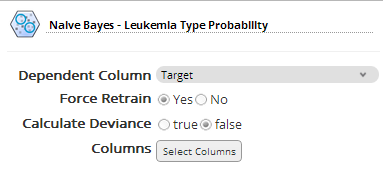
The Naive Bayes Operator is configured with the Target column as the dependent variable, as follows:
All the datasets gene columns are selected (except for the first ID column that represents the sample number).
Results
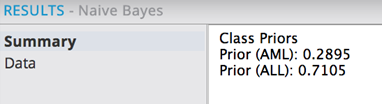
Saving and running the model produces the following results. The Summary results tab displays:
The training dataset has 28.95% occurrence of AML class of leukemia and 71.05% occurrence of ALL class of leukemia. This provides information on the historical training dataset.
Note: it is important that the priors being used to train the model are similar to the class distribution in the overall population.
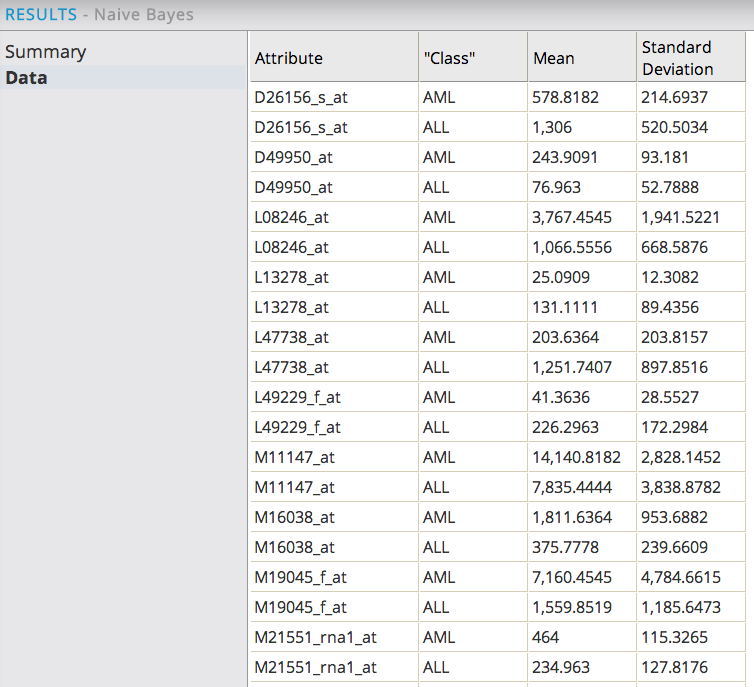
The Data results tab displays each of the top 31 predicting gene attributes along with their normal distribution curve Mean and Standard Deviation calculations.
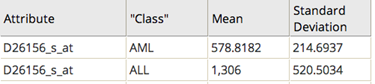
This data is helpful for understanding the measurement values that indicate the different classes of leukemia and for selecting the genes that are the most predictive of the type of Leukemia. For example, the D26156 gene attribute indicates AML class of leukemia when it has a value of 578 +/- 215, whereas it indicates ALL class of leukemia when it has a value of 1306 +/- 520.
Note: in this use case, most of the normal distribution curves for each class of leukemia are different from each other (that is, different Means and small Standard Deviations). This is because the starting dataset already reduced over 7,000 different genes down to these top 31 most predictive leukemia genes. So we expect that most of the genes are significant to the model.
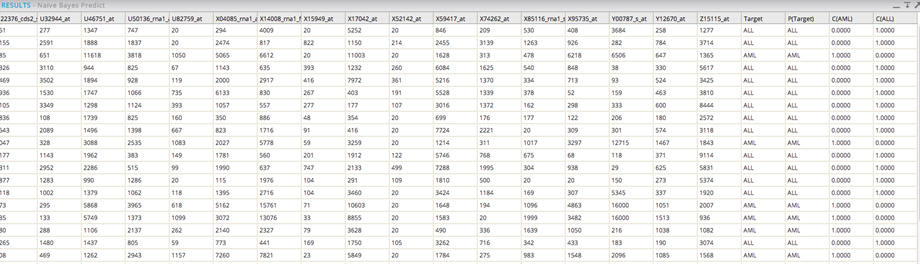
Including the Naive Bayes Predictor Operator provides additional model results:
The prediction value (0 or 1) uses a threshold assumption of > than 50% confidence that the prediction will happen.
The C(AML) column indicates the confidence that the dependent value will be AML.
The C(ALL) column indicates the confidence that the Dependent value will be ALL.
The ROC, Goodness of Fit and LiFT scoring operators should also be added to assess the model.
Summary
These results indicate the Naive Bayes model is highly predictive.
The modeler can then test the model on a different dataset than the training dataset to illustrate how the model can be used to predict leukemia types given new samples.
- Add the Leukemia Test dataset to the flow, with the same column headings but where the class (target value) of leukemia is not yet known.
- Run the following flow configuration with the test dataset connected to the Naive Bayes Predictor instead of the training dataset.
- Running the model now provides predictions for which type of leukemia each sample row contains with high confidence, illustrating the predictive power of a Naive Bayes model.